Photo courtesy of Ian McClellan
* This article has been updated. See note at the end.
Back in 2009, biophysicist Yibing Shan produced a hit movie. Though it never made it to the big screen, the 16-second video earned raves from other researchers working in the field of computational drug design.
“Mind boggling!” “New insights!” “Amazing!”
It was the first time anyone had seen such a detailed computer simulation of a drug molecule “swimming” around a target protein, seeking a site to attach and deliver its therapeutic payload. In the QuickTime snippet, one can see the protein react to the molecule by transforming shape, revealing previously invisible binding sites, known as cryptic sites.
The “Drug Swimming Movie,” as Shan casually dubbed it, revealed striking insights into what happens at the molecular level when patients receive a drug. It also dramatizes the challenges of designing drugs for proteins in a dynamic, living system.
“The idea was physics-based simulation of proteins,” says the 55-year-old Shan (MS ’98, MS ’00, and PhD ’01), who came up with the groundbreaking concept while a founding member of D. E. Shaw Research, or DESRES, a private research company where — in the early aughts — Shan contributed to the development of a pathbreaking supercomputer known as Anton. “That was very innovative.”
Now, as an independent researcher and chief computational scientist at startup BAKX Therapeutics, Shan is using the concept to model many possible drug interactions at once, combining state-of-the-art platforms with artificial intelligence and novel biology. Ultimately, this technology could lead to cures for some of medicine’s most intractable cancers and neurodegenerative disorders.
Elusive Drug Targets
For decades, a drug binding to a protein was thought of as a lock and a key, a static interaction. Shan’s swimming movies revealed a world of motion, a protein always shape-shifting — and an approaching drug molecule further influencing that shape.
Understanding the significance of Shan’s simulations — and their central role in the current computational drug discovery resurgence — demands a quick lesson in protein structures. Proteins are strings of beads made up of a sequence of chemicals known as amino acids. Attraction and repulsion between these 20 different amino acids cause the string to fold into origami-like shapes. The way the protein folds and unfolds as it interacts with other molecules determines its function in the body.
“So in theory, one can predict the structure of the protein from the amino acid sequence,” says Shan. “For a long time, people have been trying to achieve accurate predictions.”
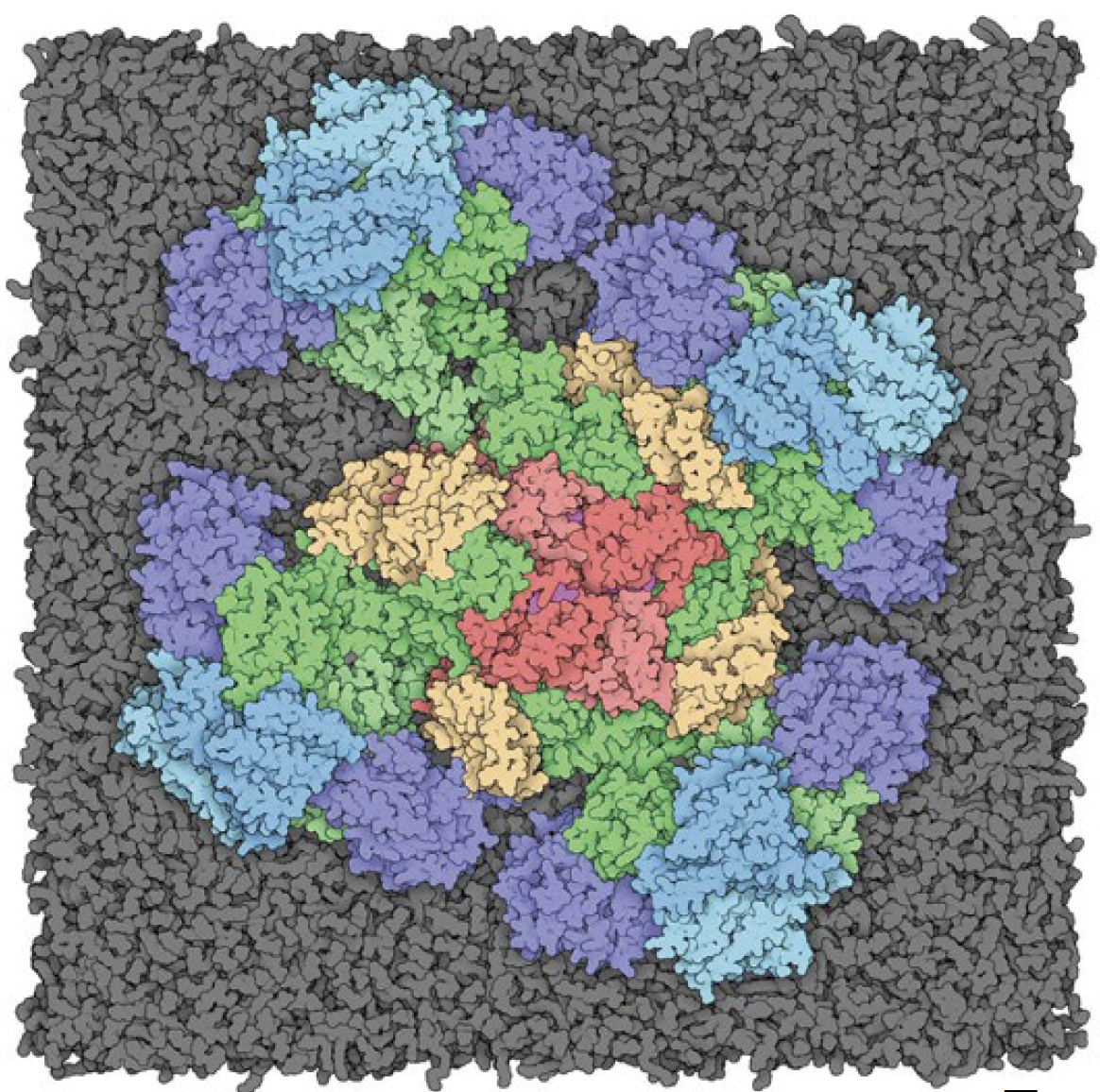
Shan is studying two proteins known to be involved in the proliferation of cancer, Ras and Raf. This model shows how the two combine in the body — it’s helpful to researchers looking for ways to disrupt the relationship between them to potentially slow and stop cancer growth. Ras protein is known as the holy grail target in cancer treatment.
Advances in brute force computing power and AI are now accelerating the field.
In the summer of 2021, for instance, DeepMind presented what commentators have called “the most important achievement in AI:” a software platform called AlphaFold that could predict the 3D structure of virtually every protein in the human body, and every protein known to science, even. Like the mapping of the human genome, it’s a triumph for researchers, sure to transform medicine and bioengineering.
But AlphaFold has its limits. It is less accurate in predicting how a protein may change shape as it moves through the body or as it interacts with, say, a drug molecule.
That’s where Shan’s swimming movie comes in. Shan’s work in DESRES and his ability to dynamically model these proteins put him on an early path toward using computer power to find human cures. In 2020, after 18 years with DESRES, he left to pursue other interests, including a venture fund that invests in health care startups seeded by an entrepreneur and friend who was fighting a form of lung cancer Shan studied. In 2021, he invested in BAKX Therapeutics, which has offices in New York and Boston, and soon after joined the firm’s leadership. Their aim is to apply what he knows to target BAX, a family of proteins that control cell death, with the goal of curing cancers.
BAX are also known as death proteins. They are the final kill switches involved in a cell’s programmed cell death, i.e. apoptosis. The company’s idea is to develop drugs that activate the cell death of cancer cells. On the flip side, BAKX also is working on inhibiting the same proteins in order to prevent cell death — useful in targeting neurodegenerative diseases. BAKX takes advantage of cloud computing to run its platform that screens molecules.
“Computational drug discovery is undergoing a renaissance,” says Loren D. Walensky, a pediatric oncologist at Dana-Farber Cancer Institute at Harvard Medical School, who is one of the BAKX co-founders and chair of its scientific board. “The limitations are really computational power, predictive power,” he says. Even a decade ago, drug development was a slow slog based on hands-on screening. “Now a lot of projects are beginning with computational screening of, in some cases, millions of molecules, done by a computer mining those structures.”
“Yibing,” Walensky adds, “is really a leader in this area.”
A 2010 article in Nature raved about Anton’s ability to simulate a protein’s 3D structure over a millisecond — a hundred-fold greater than the previous record: “Anton goes further by providing a rare, detailed glimpse into the dynamic life of a protein as it folds and unfolds, twists and wriggles.” The journal Nature Methods noted that year that Anton was “yielding new insights into protein folding and dynamics,” adding that “with Anton, the time barrier limiting the usefulness of molecular dynamics simulations has been broken.”
Shan compares the feat to a feature-length movie. “If you’re limited by the length of the movie you make, you can’t tell the whole story,” he says. Supercomputer-produced movies of drug molecule-protein interactions allow scientists to “really watch how the proteins move,” he says. “Not just a glimpse of it.”
Kevan Shokat, a cellular and molecular pharmacology professor at the University of California-San Francisco, remembers the first time he saw the “Drug Swimming Movie” in the 2000s.
“It blew my mind,” he says. “It’s a fascinating dance, just mind-boggling. It’s like a microscope for biologists. It’s really the frontier, seeing with your eyes what you had to imagine.”
Biophysics Meets Computer Science
Growing up in Shanghai, Shan says he always wanted to be a physicist, and not only because he was a good student. “As a kid, I read a lot of biographies of giants in physics,” he says of the likes of Einstein, Schrödinger and Pauli, “and I thought they were having a lot of fun. I got a naive impression that they travel around visiting each other, discussing science at parties and outdoing one another’s practical jokes.”
After graduating from the University of Shanghai for Science and Technology in 1990 with a mechanical engineering degree and working for a few years, Shan applied to graduate schools in the United States, following a friend to Drexel.
Here, he became a triple Dragon, completing two master’s degrees in physics (’98) and computer science (’99), and a doctorate in biophysics (’01).
“He was an excellent student,” says Huan-Xiang Zhou, Shan’s thesis adviser who now is a professor of chemistry and physics and the LAS Endowed Chair in the Natural Sciences at the University of Illinois at Chicago. “He was quick, and he worked hard.”
In fact, Shan approached his research with such complete concentration that the department secretary often asked him if he was upset. “No,” Shan says he responded. “Then I realized I just look intense. I was passionate about it.
“I’m still not a big smiler,” he adds.
Shan’s thesis focused on modeling protein-protein interactions that form complexes, by figuring out where specifically the proteins bind. The work, Zhou says, predates DeepMind’s AlphaFold by nearly two decades. (As it happens, John Jumper, MD, who led the AlphaFold team in DeepMind, took a detour after his undergraduate study and worked closely with Shan in DESRES for a couple of years.)
Like AlphaFold, Shan used a neural network trained on a database to make actual predictions about the protein-to-protein interface.
“That was machine learning,” Shan says. “That turned out to be important, although I certainly didn’t quite see the rage it enjoys today. Even then, the purpose was drug discovery.”
Zhou says he did the “dirty work” of building the database manually, while Shan did the “intelligent work” of designing and programming the neural network. The resulting 2001 paper — “Prediction of Protein Interaction Sites from Sequence Profile and Residue Neighbor List” in PROTEINS: Structure, Function, and Bioinformatics — was highly cited, he says.
“It worked out remarkably well,” Zhou says, “so much better than I ever anticipated. It really opened a new area.”
After a stint at a startup, Shan “lucked into” an interview at DESRES in 2002, he says. A wide-ranging three-and-half-hour conversation with David Shaw led to an offer that made him among the company’s first hires.
In those early years at DESRES, Shan and colleagues read and explored possible projects to pitch to Shaw. They settled on developing a supercomputer specialized in molecular dynamics simulations, which is now known as Anton. It was “extremely exciting,” he says. “You learn a lot about how to conceptualize an ambitious project.”
His deep dive led to the development of the Gaussian Split Ewald algorithm, which allows fast electrostatic calculations and has proven to be a key design feature of Anton. In 2009, he and colleagues won the Association for Computing Machinery’s Gordon Bell Prize for the millisecond Anton simulation.
After the initial supercomputer was up and running, Shan shifted to applications. One, of course, was the drug-molecule-protein interaction captured visually by his swimming movie. In a 2014 talk at the University of Washington, David Shaw noted his initial skepticism about Shan’s idea of using Anton to simulate the interaction between a small drug molecule and a target protein known to be involved in leukemia.
“If he had asked me first,” Shaw said in the talk, “I would have said, ‘Yibing, that’s a great idea, but realistically what are the odds it’s going to work? You’ll use up Anton time, and you’re busy working on stuff that’s higher priority.
“The only thing is,” he continued, “that Yibing was right, and it actually worked.”
Numerous publications flowed from Shan’s simulations, including two high-profile papers in the journal Cell in 2012 and 2013. In the first paper, Shan reported modeling the mutated structure of Epidermal Growth Factor Receptor (EGFR), a protein involved in cell proliferation and a drug target for lung and other cancers. The research uncovered the mechanism by which the EGFR mutations promote cancer growth. The second paper focused on the structure of EGFR in a realistic membrane environment.
One of his last projects at DESRES modeled the cancer drug target K-Ras, a protein considered the “holy grail” in cancer drug discovery, Shan says. K-Ras instructs a cell to grow and divide or take on specialized functions, but when it’s overactive, cancer proliferates. The work, published in 2021, relied heavily on Anton, he says, “to build such a comprehensive and huge structure.”
Shaw described Shan as a “gifted researcher who’s already … made important contributions involving both new technologies and methodologies for computational chemistry research and advances in the scientific understanding of molecular mechanisms underlying biologically and pharmaceutically relevant processes.”
Shan’s past and future work will wind up making a real difference in the lives of patients, Shaw predicts.
Ushering In a New Era
Computational drug discovery and simulations of drug protein interactions can seem like a magic bullet for cancer or any number of other hard-to-target conditions. So far, the technology has not fully realized its promise, though startups such as BAKX are hard at work to change that outcome.
“The whole approach is really showing its worth in terms of being able to actually initiate the process of drug discovery,” Walensky says, “and the validation, of course, comes from rigorous experimentation.”
According to BAKX CEO and founder Sree Kant, the trick is the right combination of ingredients — and a prime one is the swimming simulation guy, himself.
“He’s well known as one of the godfathers of computational drug discovery,” Kant says, adding that Shan brings “huge amounts of curiosity combined with objective honesty. You need to be curious, but you also need to be very critical of what you’re doing.”
The BAKX platform, built on Shan’s body of research, screens small molecules at a massive scale, validates site-specific binding and narrows down a universe of variables to find optimal drug candidates, Kant says. The startup employs both computational and bench scientists, he adds.
“Previously, biology has been very accidental,” Kant says. “We don’t understand the entire human body, how everything works.” At the same time, the trove of data based on biological observations and computing power is growing. “The thought has been that if you harness enough understanding of data, you can be more predictive.”
Accurate predictions could reduce the timeline and huge costs of drug discovery, even as they reveal new and more precise targets for challenging problems. BAKX is particularly interested in so-called undruggable targets, which have drug-binding sites that are cryptic, meaning flat and featureless or changeable.
Kant says the company is zeroing in on molecules that target the apoptotic proteins and is partnering with Ipsen Biopharmaceuticals to initiate clinical trials in the near future.
For Shan, the realization of what is truly at stake for BAKX and for this field of research came after an encounter at an academic conference in 2015. After his talk, Shan was approached by a young woman whose husband was suffering from lung cancer.
“They have three young children, and she asked a bunch of questions about what was the best approach of treatment, about what they could do,” he says. “I didn’t have very good answers. I was so crushed.
“I think that’s what this game is about,” he continues. “This is about really helping people. We’re so much closer to showing this technology helps drug discovery. It’s a very delicate thing to get it right. Eventually, we will.”
* Drexel Magazine was surprised to learn, soon after publication, that BAKX will soon be ceasing operations. Rather than remove the story, which reflects information available as of publication, we’ve decided to preserve it as a profile of an alumnus. Startups are inherently risky enterprises, and we wish Shan well in his future pursuits. He told us the following, soon after publication: “While I am disappointed that BAKX will be ceasing operations, due to the challenging funding environment and a number of other factors, I will continue to invest in other companies and to develop and apply computational science and AI-based discovery approaches through my venture company, AB Magnitude Ventures.”